Tree-based inference and hyperparameter optimization
Lecture 19
Cornell University
INFO 5001 - Fall 2025
November 4, 2025
Announcements
Announcements
TODO
Learning objectives
- Define tree-based inference
- Identify hyperparameters for machine learning models
- Fit decision tree and random forest models
- Tune hyperparameters using a grid search
- Identify the best model and finalize the workflow
Application exercise
ae-17
Note
- Go to the course GitHub org and find your
ae-17(repo name will be suffixed with your GitHub name). - Clone the repo in Positron, run
renv::restore()to install the required packages, open the Quarto document in the repo, and follow along and complete the exercises. - Render, commit, and push your edits by the AE deadline – end of the day
Decision trees
Decision Trees
To predict the outcome of a new data point:
- Uses rules learned from splits
- Each split maximizes information gain
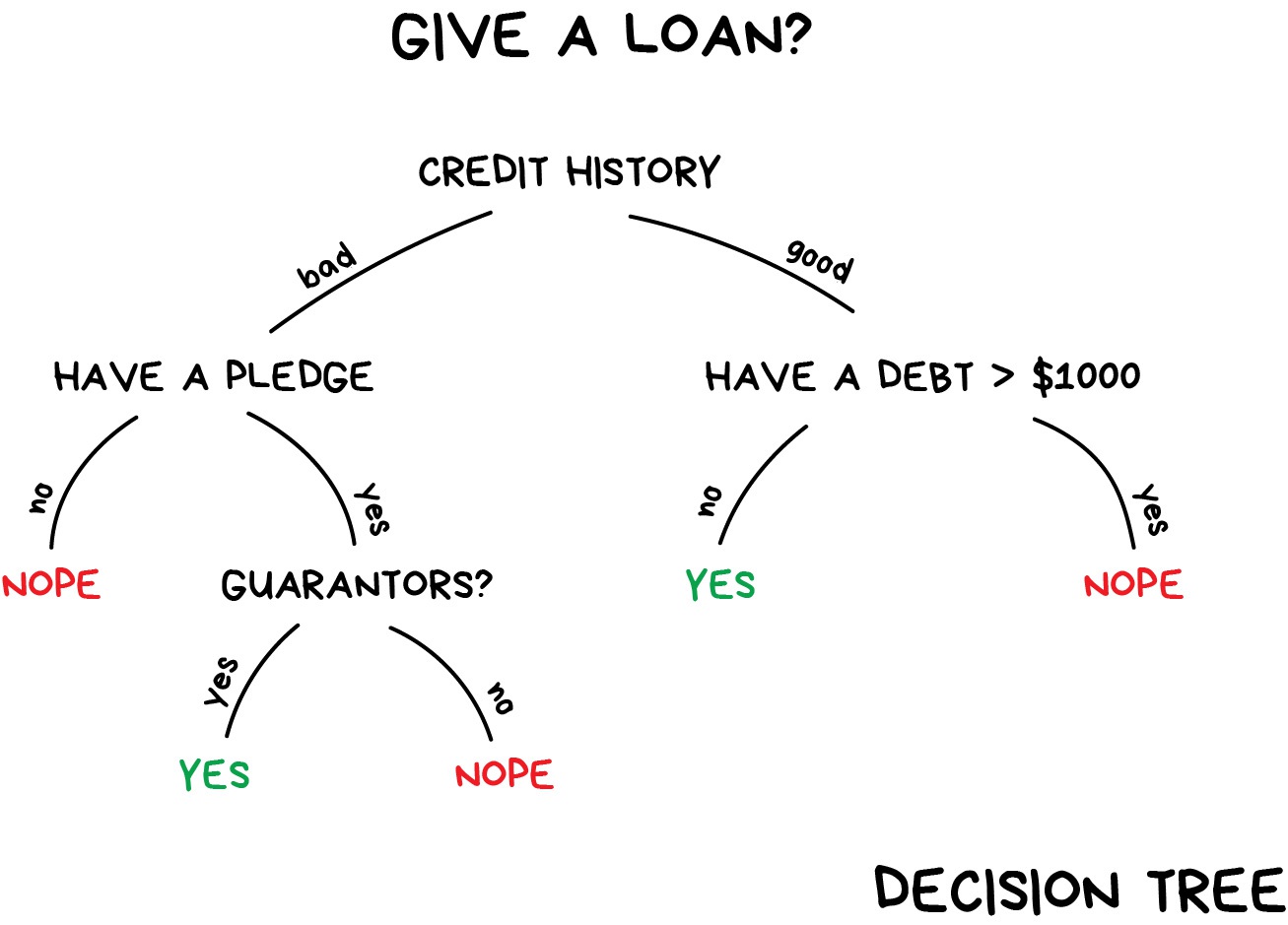
Image credit: Machine Learning for Everyone


Quiz
How do we assess predictions here?
Root Mean Squared Error (RMSE)

Uses of decision trees
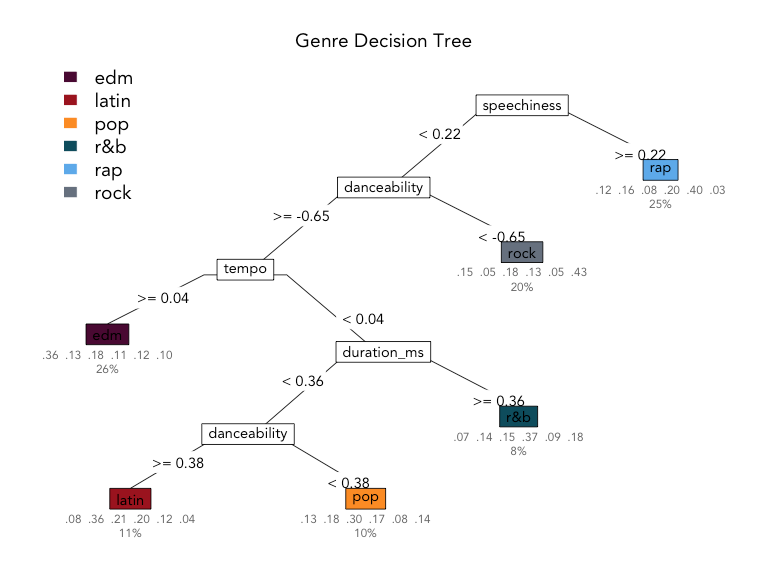
Image credit: Kaylin Pavlik


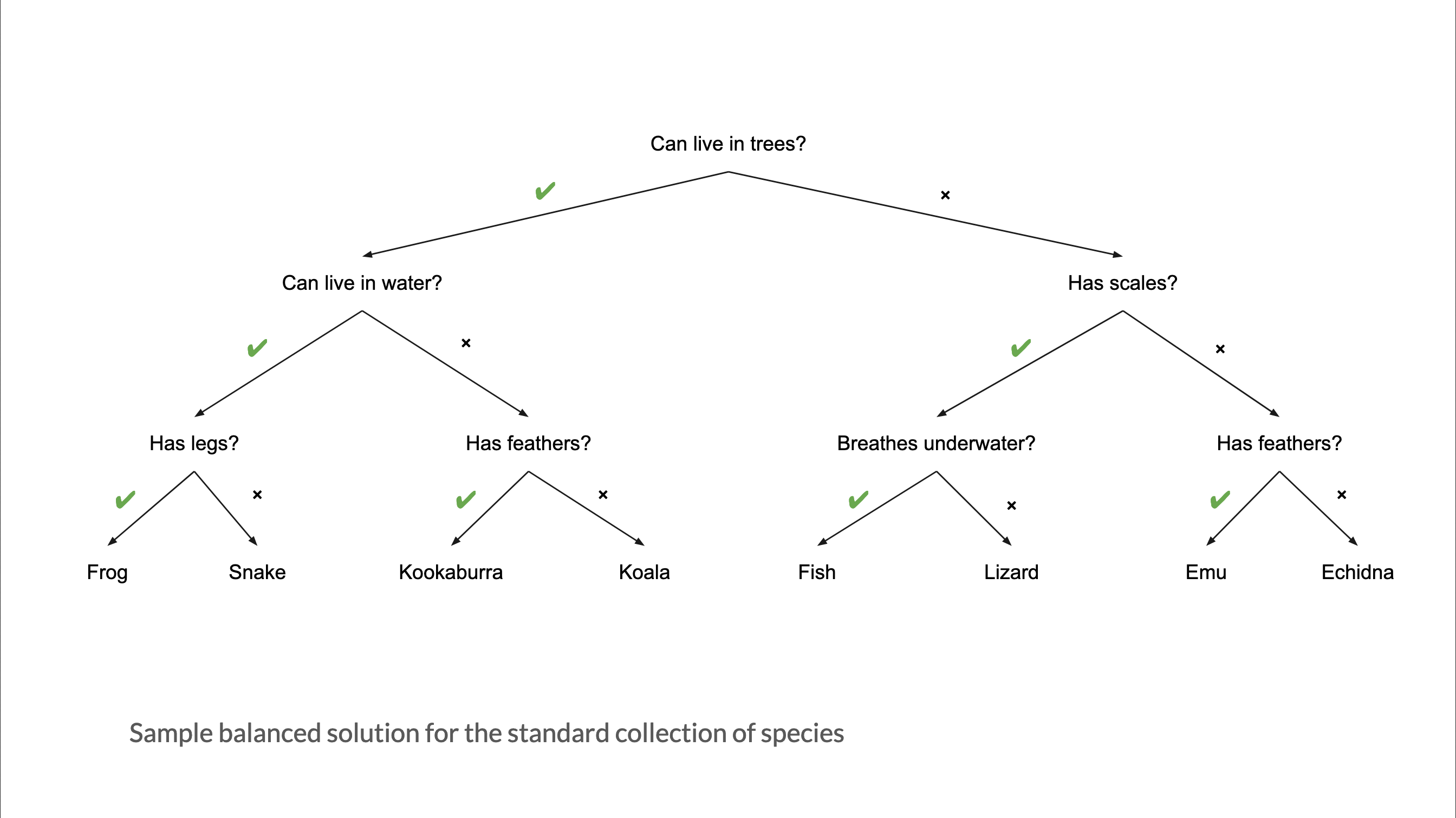
What makes a good guesser?
High information gain per question (can it fly?)
Clear features (feathers vs. is it “small”?)
Order of features matters

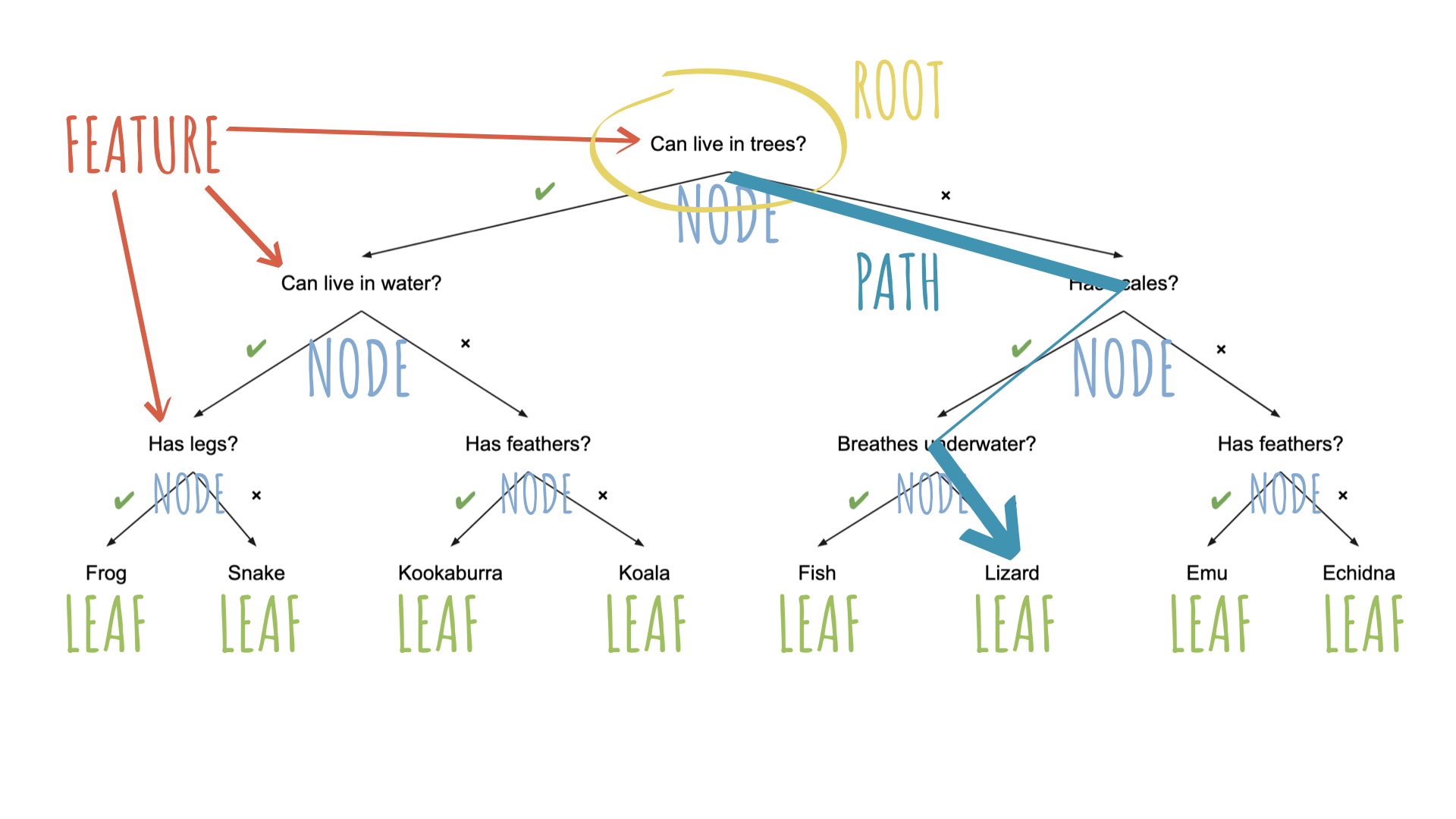
Source: Australian Computing Academy
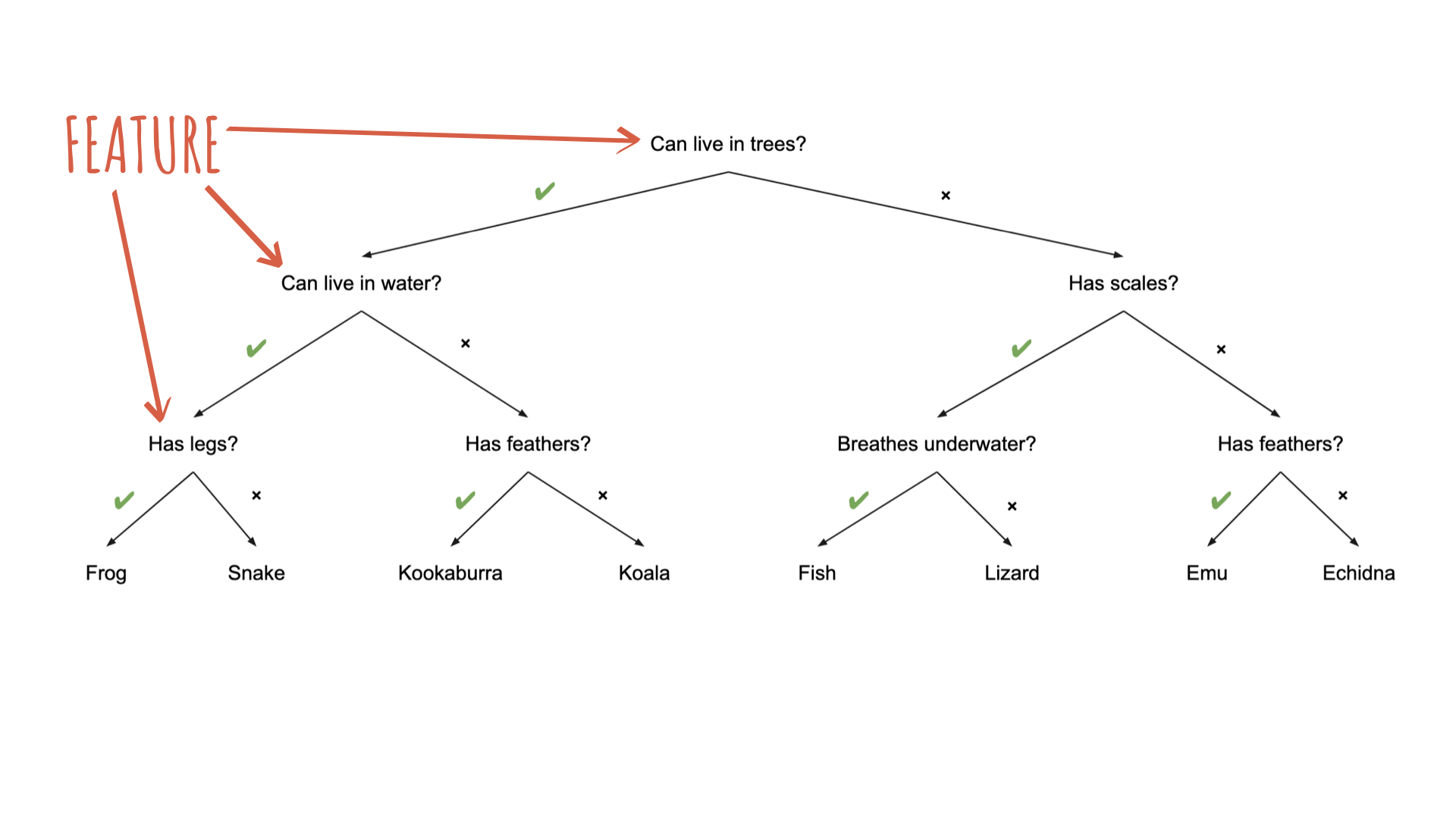
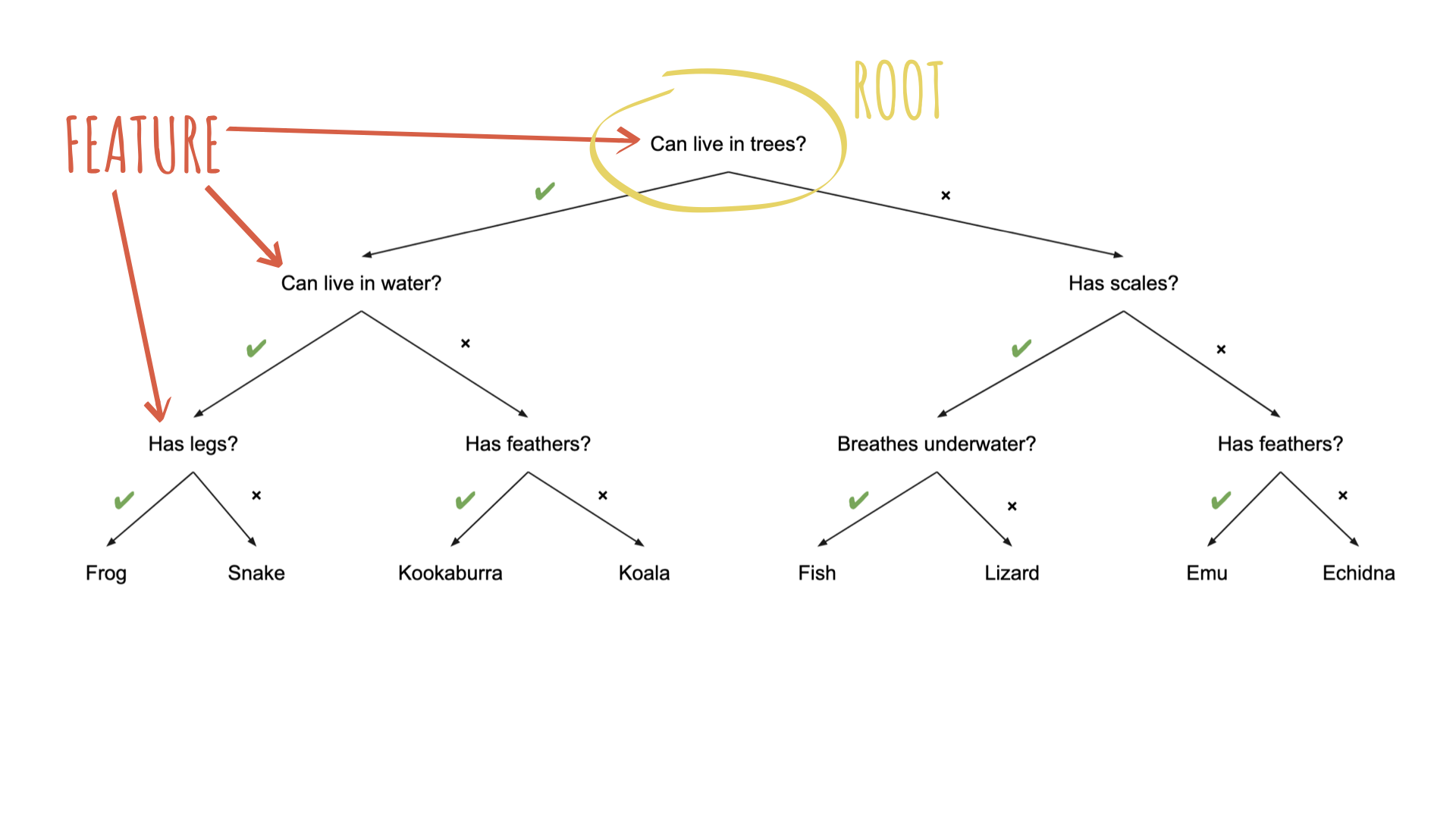
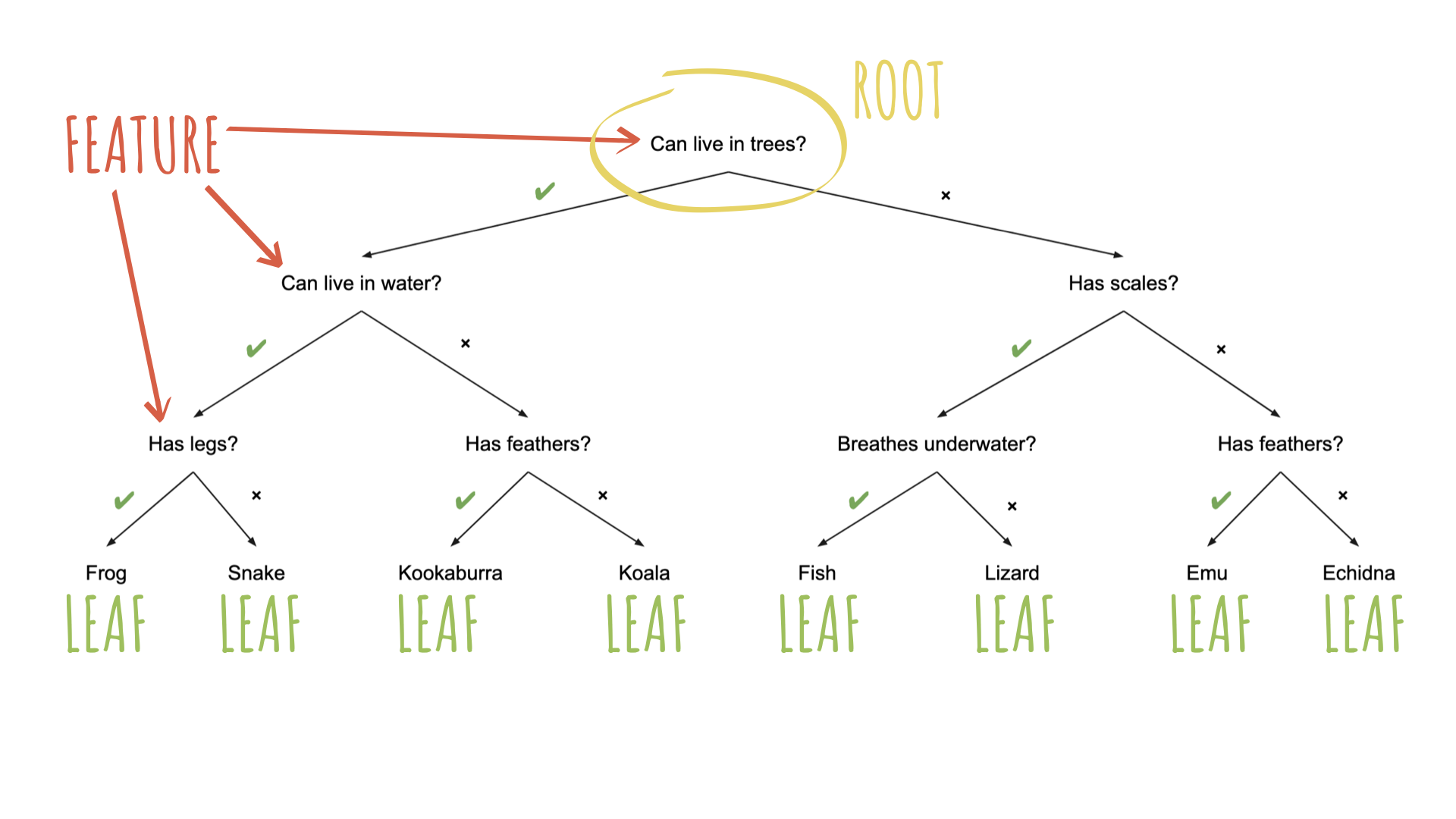
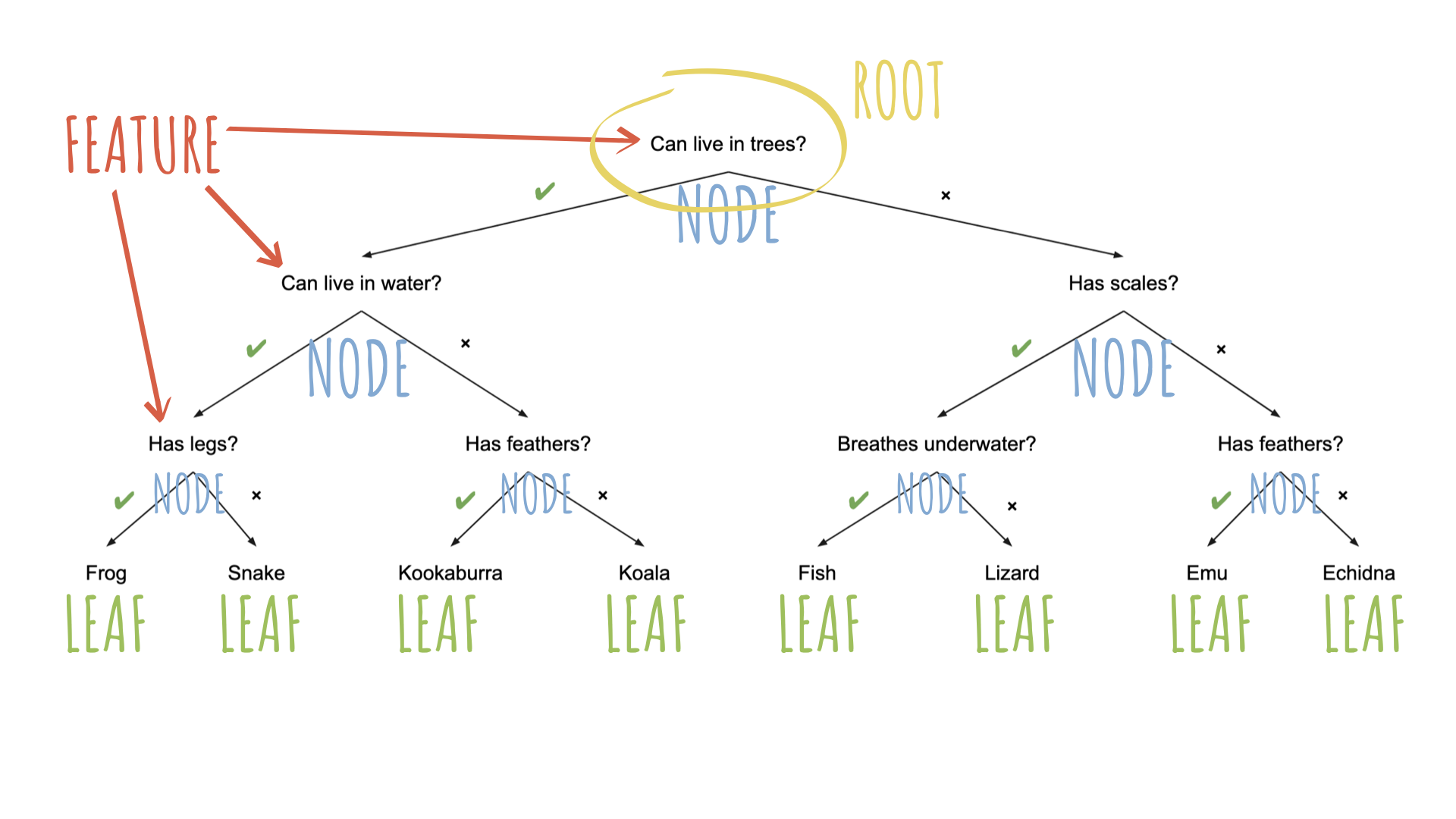
To specify a model with {parsnip}
1. Pick a model + engine
2. Set the mode (if needed)
To specify a decision tree model with {parsnip}

nn children chi non cover
2 children [.77 .23] when average_daily_rate >= 130 37%
3 none [.35 .65] when average_daily_rate < 130 63%

⌨️ Fit a decision tree with cross-validation
Here is our very-vanilla parsnip model specification for a decision tree (also in your qmd)…
And a workflow:
For decision trees, no recipe really required 🎉
⌨️ Fit a decision tree with cross-validation
Instructions
Fill in the blanks to return the accuracy and ROC AUC for this model using 10-fold cross-validation.
02:00
# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.786 10 0.00706 pre0_mod0_post0
2 brier_class binary 0.156 10 0.00417 pre0_mod0_post0
3 roc_auc binary 0.833 10 0.00840 pre0_mod0_post0Model arguments
args()
Print the arguments (hyperparameters) for a {parsnip} model specification.
decision_tree()
Specifies a decision tree model
either mode works!
decision_tree()
Specifies a decision tree model
set_args()
Change the arguments for a {parsnip} model specification.
tree_depth
Cap the maximum tree depth.
A method to stop the tree early. Used to prevent overfitting.


min_n
Set minimum n to split at any node.
Another early stopping method. Used to prevent overfitting.
Quiz
What value of min_n would lead to the most overfit tree?
min_n = 1
Recap: early stopping
| {parsnip} arg | rpart arg |
default | overfit? |
|---|---|---|---|
tree_depth |
maxdepth |
30 | ⬆️ |
min_n |
minsplit |
20 | ⬇️ |
cost_complexity
Adds a cost or penalty to error rates of more complex trees.
A way to prune a tree. Used to prevent overfitting.
Closer to zero ➡️ larger trees.
Higher penalty ➡️ smaller trees.

Consider the bonsai

Small pot
Strong shears
Consider the bonsai

Small potEarly stoppingStrong shearsPruning
Recap: early stopping & pruning
| {parsnip} arg | rpart arg |
default | overfit? |
|---|---|---|---|
tree_depth |
maxdepth |
30 | ⬆️ |
min_n |
minsplit |
20 | ⬇️ |
cost_complexity |
cp |
.01 | ⬇️ |







Axiom
There is an inverse relationship between model accuracy and model interpretability.
Random forests

Ensemble methods
Ensemble methods combine many simple “building block” models in order to obtain a single and potentially very powerful model.
- Individual models are typically weak learners - low accuracy on their own
- Combining a set of weak learners can create a strong learner with high accuracy by reducing bias and variance
More info: Introduction to Statistical Learning with R
Bagging
Decision trees suffer from high variance - small changes in the data can lead to very different trees
Bootstrap aggregation (or bagging) reduces the variance of a model by averaging the predictions of many models trained on different samples of the data.
Random forests
To further improve performance over bagged trees, random forests introduce additional randomness in the model-building process to reduce the correlation across the trees.
- Random feature selection: At each split, only a random subset of features are considered
- Makes each individual model simpler (and “dumber”), but improves the overall performance of the forest
rand_forest()
Specifies a random forest model
either mode works!
rand_forest()
Specifies a random forest model
⌨️ Fit a random forest
Instructions
Create a new parsnip model called rf_mod, which will learn an ensemble of classification trees from our training data using the {ranger} engine. Update your tree_wf with this new model.
Fit your workflow with 10-fold cross-validation and compare the ROC AUC of the random forest to your single decision tree model — which predicts the assessment set better?
Hint: you’ll need https://www.tidymodels.org/find/parsnip/
04:00
# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.827 10 0.00660 pre0_mod0_post0
2 brier_class binary 0.122 10 0.00226 pre0_mod0_post0
3 roc_auc binary 0.913 10 0.00387 pre0_mod0_post0mtry
The number of predictors that will be randomly sampled at each split when creating the tree models.
{ranger} default = floor(sqrt(num_predictors))
Single decision tree
tree_mod <- decision_tree(engine = "rpart") |>
set_mode("classification")
tree_wf <- workflow() |>
add_formula(children ~ .) |>
add_model(tree_mod)
set.seed(100)
tree_res <- tree_wf |>
fit_resamples(
resamples = hotels_folds,
control = control_resamples(save_pred = TRUE)
)
tree_res |>
collect_metrics()# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.786 10 0.00706 pre0_mod0_post0
2 brier_class binary 0.156 10 0.00417 pre0_mod0_post0
3 roc_auc binary 0.833 10 0.00840 pre0_mod0_post0A random forest of trees
# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.827 10 0.00660 pre0_mod0_post0
2 brier_class binary 0.122 10 0.00226 pre0_mod0_post0
3 roc_auc binary 0.913 10 0.00387 pre0_mod0_post0Different mtry values
mtry = 5
# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.830 10 0.00567 pre0_mod0_post0
2 brier_class binary 0.121 10 0.00242 pre0_mod0_post0
3 roc_auc binary 0.913 10 0.00405 pre0_mod0_post0mtry = 12
# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.828 10 0.00473 pre0_mod0_post0
2 brier_class binary 0.122 10 0.00257 pre0_mod0_post0
3 roc_auc binary 0.909 10 0.00414 pre0_mod0_post0mtry = 21
# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.822 10 0.00597 pre0_mod0_post0
2 brier_class binary 0.124 10 0.00272 pre0_mod0_post0
3 roc_auc binary 0.906 10 0.00411 pre0_mod0_post0🎶 Fitting and tuning models with {tune}
Hyperparameters
- Model parameters/tuning parameters
- Some parameters can be estimated directly from the training data
- Regression coefficients
- Split points
- Some parameters (hyperparameters or tuning parameters) must be specified ahead of time
- Number of trees in a random forest
- Number of neighbors in k-nearest neighbors
- Number of layers in a neural network
- Learning rate in a gradient boosting machine
tune()
A placeholder for hyper-parameters to be “tuned”
tune_grid()
A version of fit_resamples() that performs a grid search for the best combination of tuned hyper-parameters.
tune_grid()
A version of fit_resamples() that performs a grid search for the best combination of tuned hyper-parameters.
tune_grid()
A version of fit_resamples() that performs a grid search for the best combination of tuned hyper-parameters.
⌨️ Tune a random forest
Here’s our random forest model plus workflow to work with.
⌨️ Tune a random forest
Here is the output from fit_resamples()…
# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.827 10 0.00660 pre0_mod0_post0
2 brier_class binary 0.122 10 0.00226 pre0_mod0_post0
3 roc_auc binary 0.913 10 0.00387 pre0_mod0_post0⌨️ Tune a random forest
Instructions
Edit the random forest model to tune the mtry and min_n hyperparameters.
Update your workflow to use the tuned model.
Then use tune_grid() to find the best combination of hyper-parameters to maximize roc_auc; let {tune} set up the grid for you.
How does it compare to the average ROC AUC across folds from fit_resamples()?
05:00
# A tibble: 30 × 8
mtry min_n .metric .estimator mean n std_err
<int> <int> <chr> <chr> <dbl> <int> <dbl>
1 1 14 accuracy binary 0.787 10 0.00519
2 1 14 brier_class binary 0.172 10 0.00157
3 1 14 roc_auc binary 0.880 10 0.00422
4 3 31 accuracy binary 0.825 10 0.00661
5 3 31 brier_class binary 0.128 10 0.00211
6 3 31 roc_auc binary 0.908 10 0.00395
7 5 2 accuracy binary 0.829 10 0.00564
8 5 2 brier_class binary 0.121 10 0.00254
9 5 2 roc_auc binary 0.913 10 0.00423
10 7 18 accuracy binary 0.831 10 0.00573
# ℹ 20 more rows
# ℹ 1 more variable: .config <chr># A tibble: 300 × 7
id mtry min_n .metric .estimator .estimate .config
<chr> <int> <int> <chr> <chr> <dbl> <chr>
1 Fold01 1 14 accuracy binary 0.797 pre0_m…
2 Fold01 1 14 roc_auc binary 0.895 pre0_m…
3 Fold01 1 14 brier_cl… binary 0.166 pre0_m…
4 Fold02 1 14 accuracy binary 0.783 pre0_m…
5 Fold02 1 14 roc_auc binary 0.876 pre0_m…
6 Fold02 1 14 brier_cl… binary 0.173 pre0_m…
7 Fold03 1 14 accuracy binary 0.808 pre0_m…
8 Fold03 1 14 roc_auc binary 0.889 pre0_m…
9 Fold03 1 14 brier_cl… binary 0.169 pre0_m…
10 Fold04 1 14 accuracy binary 0.781 pre0_m…
# ℹ 290 more rowstune_grid()
A version of fit_resamples() that performs a grid search for the best combination of tuned hyper-parameters.
expand_grid()
Takes one or more vectors, and returns a data frame holding all combinations of their values.
show_best()
Shows the n most optimum combinations of hyper-parameters
# A tibble: 5 × 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 5 2 roc_auc binary 0.913 10 0.00423 pre0_m…
2 7 18 roc_auc binary 0.912 10 0.00373 pre0_m…
3 12 6 roc_auc binary 0.910 10 0.00411 pre0_m…
4 9 35 roc_auc binary 0.908 10 0.00389 pre0_m…
5 3 31 roc_auc binary 0.908 10 0.00395 pre0_m…autoplot()
Quickly visualize tuning results

fit_best()
- Replaces
tune()placeholders in a model/recipe/workflow with a set of hyper-parameter values - Fits a model using the entire training set

We are ready to touch the jewels…
The testing set!
⌨️ Evaluate on the test set
Instructions
Use fit_best() to take the best combination of hyper-parameters from rf_results and use them to predict the test set.
How does our actual test ROC AUC compare to our cross-validated estimate?
05:00
# A tibble: 1 × 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 5 2 roc_auc binary 0.913 10 0.00423 pre0_m…# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 roc_auc binary 0.907ROC curve

The entire process
The set-up
The tune-up
# here comes the actual ML bits…
# pick model to tune
rf_tuner <- rand_forest(
engine = "ranger",
mtry = tune(),
min_n = tune()
) |>
set_mode("classification")
rf_wf <- workflow() |>
add_formula(children ~ .) |>
add_model(rf_tuner)
rf_results <- rf_wf |>
tune_grid(
resamples = hotels_folds,
control = control_grid(
save_pred = TRUE,
save_workflow = TRUE
)
)Quick check-in…
# A tibble: 10 × 4
# Groups: .config, mtry [10]
.config mtry min_n folds
<chr> <int> <int> <int>
1 pre0_mod01_post0 1 14 10
2 pre0_mod02_post0 3 31 10
3 pre0_mod03_post0 5 2 10
4 pre0_mod04_post0 7 18 10
5 pre0_mod05_post0 9 35 10
6 pre0_mod06_post0 12 6 10
7 pre0_mod07_post0 14 23 10
8 pre0_mod08_post0 16 40 10
9 pre0_mod09_post0 18 10 10
10 pre0_mod10_post0 21 27 10The match up!
# A tibble: 5 × 8
mtry min_n .metric .estimator mean n
<int> <int> <chr> <chr> <dbl> <int>
1 5 2 roc_auc binary 0.914 10
2 7 18 roc_auc binary 0.911 10
3 12 6 roc_auc binary 0.910 10
4 3 31 roc_auc binary 0.909 10
5 9 35 roc_auc binary 0.908 10
# ℹ 2 more variables: std_err <dbl>,
# .config <chr># A tibble: 1 × 3
mtry min_n .config
<int> <int> <chr>
1 5 2 pre0_mod03_post0
The wrap-up
══ Workflow [trained] ══════════════════════════════════════════════════════════════════════════════
Preprocessor: Formula
Model: rand_forest()
── Preprocessor ────────────────────────────────────────────────────────────────────────────────────
children ~ .
── Model ───────────────────────────────────────────────────────────────────────────────────────────
Ranger result
Call:
ranger::ranger(x = maybe_data_frame(x), y = y, mtry = min_cols(~5L, x), min.node.size = min_rows(~2L, x), num.threads = 1, verbose = FALSE, seed = sample.int(10^5, 1), probability = TRUE)
Type: Probability estimation
Number of trees: 500
Sample size: 3600
Number of independent variables: 21
Mtry: 5
Target node size: 2
Variable importance mode: none
Splitrule: gini
OOB prediction error (Brier s.): 0.119472 Wrap up
Recap
- Decision trees are a nonlinear, naturally interactive model for classification and regression tasks
- Simple decision trees can be easily interpreted, but also less accurate
- Random forests are ensembles of decision trees used to aggregate predictions for improved performance
- Models with hyperparameters require tuning in order to achieve optimal performance
- Once the optimal model is achieved via cross-validation, fit the model a final time using the entire training set and evaluate its performance using the test set
Acknowledgments
- Materials derived from Tidymodels, Virtually by Allison Hill and licensed under a Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA) License.
- Dataset and some modeling steps derived from A predictive modeling case study and licensed under a Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA) License.
