tree_mod <- decision_tree(engine = "rpart") |>
set_mode("classification")
tree_wf <- workflow() |>
add_formula(children ~ .) |>
add_model(tree_mod)Tune better models to predict children in hotel bookings
Suggested answers
Your Turn 1
Fill in the blanks to return the accuracy and ROC AUC for this model using 10-fold cross-validation.
Fill in the blanks to return the accuracy and ROC AUC for this model using 10-fold cross-validation.
set.seed(100)
______ |>
______(resamples = hotels_folds) |>
______Answer:
set.seed(100)
tree_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.773 10 0.00567 Preprocessor1_Model1
2 brier_class binary 0.158 10 0.00322 Preprocessor1_Model1
3 roc_auc binary 0.832 10 0.00672 Preprocessor1_Model1Your Turn 2
Create a new parsnip model called rf_mod, which will learn an ensemble of classification trees from our training data using the ranger package. Update your tree_wf with this new model.
Fit your workflow with 10-fold cross-validation and compare the ROC AUC of the random forest to your single decision tree model — which predicts the test set better?
Hint: you’ll need https://www.tidymodels.org/find/parsnip/
# model
rf_mod <- _____ |>
_____("ranger") |>
_____("classification")
# workflow
rf_wf <- tree_wf |>
update_model(_____)
# fit with cross-validation
set.seed(100)
_____ |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()Answer:
# model
rf_mod <- rand_forest(engine = "ranger") |>
set_mode("classification")
# workflow
rf_wf <- tree_wf |>
update_model(rf_mod)
# fit with cross-validation
set.seed(100)
rf_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.829 10 0.00332 Preprocessor1_Model1
2 brier_class binary 0.123 10 0.00176 Preprocessor1_Model1
3 roc_auc binary 0.912 10 0.00320 Preprocessor1_Model1Your Turn 3
Challenge: Fit 3 more random forest models, each using 5, 12, and 21 variables at each split. Update your rf_wf with each new model. Which value maximizes the area under the ROC curve?
rf5_mod <- rf_mod |>
set_args(mtry = 5)
rf12_mod <- rf_mod |>
set_args(mtry = 12)
rf21_mod <- rf_mod |>
set_args(mtry = 21) Do this for each model above:
_____ <- rf_wf |>
update_model(_____)
set.seed(100)
_____ |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()Answer:
# 5
rf5_wf <- rf_wf |>
update_model(rf5_mod)
set.seed(100)
rf5_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.829 10 0.00376 Preprocessor1_Model1
2 brier_class binary 0.122 10 0.00176 Preprocessor1_Model1
3 roc_auc binary 0.912 10 0.00305 Preprocessor1_Model1# 12
rf12_wf <- rf_wf |>
update_model(rf12_mod)
set.seed(100)
rf12_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.831 10 0.00414 Preprocessor1_Model1
2 brier_class binary 0.123 10 0.00239 Preprocessor1_Model1
3 roc_auc binary 0.908 10 0.00418 Preprocessor1_Model1# 21
rf21_wf <- rf_wf |>
update_model(rf21_mod)
set.seed(100)
rf21_wf |>
fit_resamples(resamples = hotels_folds) |>
collect_metrics()# A tibble: 3 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 accuracy binary 0.827 10 0.00382 Preprocessor1_Model1
2 brier_class binary 0.125 10 0.00256 Preprocessor1_Model1
3 roc_auc binary 0.905 10 0.00438 Preprocessor1_Model1Your Turn 4
Edit the random forest model to tune the mtry and min_n hyper-parameters; call the new model spec rf_tuner.
Update your workflow to use the tuned model.
Then use tune_grid() to find the best combination of hyper-parameters to maximize roc_auc; let tune set up the grid for you.
How does it compare to the average ROC AUC across folds from fit_resamples()?
rf_mod <- rand_forest(engine = "ranger") |>
set_mode("classification")
rf_wf <- workflow() |>
add_formula(children ~ .) |>
add_model(rf_mod)
set.seed(100) # Important!
rf_results <- rf_wf |>
fit_resamples(resamples = hotels_folds,
metrics = metric_set(roc_auc),
# change me to control_grid() with tune_grid
control = control_resamples(save_workflow = TRUE))
rf_results |>
collect_metrics()# A tibble: 1 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 roc_auc binary 0.912 10 0.00320 Preprocessor1_Model1Answer:
rf_tuner <- rand_forest(
engine = "ranger",
mtry = tune(),
min_n = tune()
) |>
set_mode("classification")
rf_wf <- rf_wf |>
update_model(rf_tuner)
set.seed(100) # Important!
rf_results <- rf_wf |>
tune_grid(resamples = hotels_folds,
control = control_grid(save_workflow = TRUE))i Creating pre-processing data to finalize unknown parameter: mtryYour Turn 5
Use fit_best() to take the best combination of hyper-parameters from rf_results and use them to predict the test set.
How does our actual test ROC AUC compare to our cross-validated estimate?
hotels_best <- fit_best(rf_results)
# cross validated ROC AUC
rf_results |>
show_best(metric = "roc_auc", n = 5)# A tibble: 5 × 8
mtry min_n .metric .estimator mean n std_err .config
<int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 3 15 roc_auc binary 0.910 10 0.00283 Preprocessor1_Model07
2 8 20 roc_auc binary 0.909 10 0.00376 Preprocessor1_Model10
3 7 36 roc_auc binary 0.908 10 0.00372 Preprocessor1_Model02
4 9 28 roc_auc binary 0.907 10 0.00381 Preprocessor1_Model01
5 12 21 roc_auc binary 0.907 10 0.00430 Preprocessor1_Model03# test set ROC AUC
augment(hotels_best, new_data = hotels_test) |>
roc_auc(truth = children, .pred_children)# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
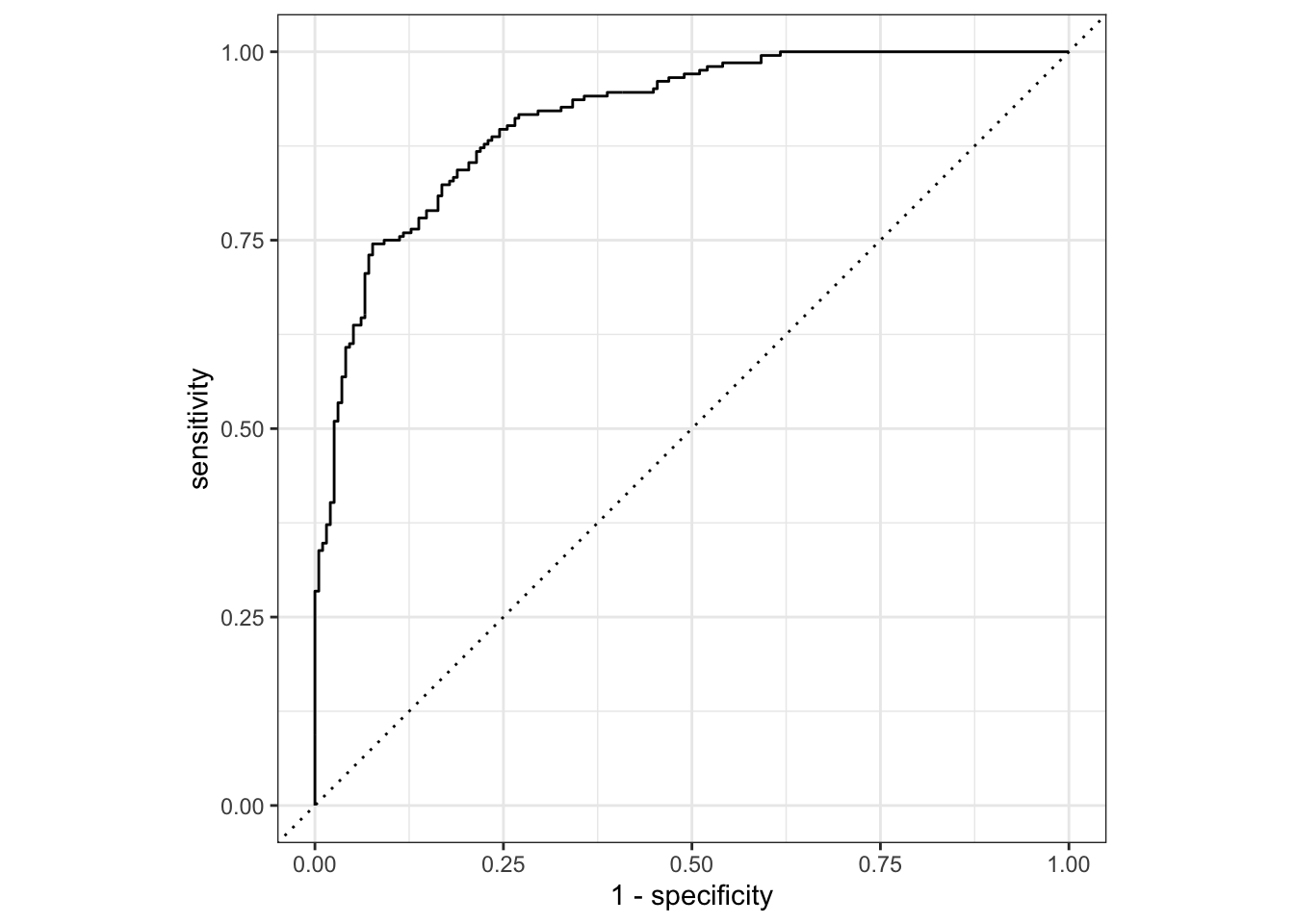
1 roc_auc binary 0.913# test set ROC curve
augment(hotels_best, new_data = hotels_test) |>
roc_curve(truth = children, .pred_children) |>
autoplot()Acknowledgments
- Materials derived from Tidymodels, Virtually: An Introduction to Machine Learning with Tidymodels by Allison Hill.
- Dataset and some modeling steps derived from A predictive modeling case study and licensed under a Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA) License.
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.4.1 (2024-06-14)
os macOS Sonoma 14.6.1
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz America/New_York
date 2024-11-12
pandoc 3.4 @ /usr/local/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
! package * version date (UTC) lib source
P backports 1.5.0 2024-05-23 [?] CRAN (R 4.4.0)
P bit 4.0.5 2022-11-15 [?] CRAN (R 4.3.0)
P bit64 4.0.5 2020-08-30 [?] CRAN (R 4.3.0)
P broom * 1.0.6 2024-05-17 [?] CRAN (R 4.4.0)
P class 7.3-22 2023-05-03 [?] CRAN (R 4.4.0)
cli 3.6.3 2024-06-21 [1] RSPM (R 4.4.0)
P codetools 0.2-20 2024-03-31 [?] CRAN (R 4.4.1)
P colorspace 2.1-0 2023-01-23 [?] CRAN (R 4.3.0)
P crayon 1.5.3 2024-06-20 [?] CRAN (R 4.4.0)
P data.table 1.15.4 2024-03-30 [?] CRAN (R 4.3.1)
P dials * 1.2.1 2024-02-22 [?] CRAN (R 4.3.1)
P DiceDesign 1.10 2023-12-07 [?] CRAN (R 4.3.1)
P digest 0.6.35 2024-03-11 [?] CRAN (R 4.3.1)
P dplyr * 1.1.4 2023-11-17 [?] CRAN (R 4.3.1)
P evaluate 0.24.0 2024-06-10 [?] CRAN (R 4.4.0)
P fansi 1.0.6 2023-12-08 [?] CRAN (R 4.3.1)
P farver 2.1.2 2024-05-13 [?] CRAN (R 4.3.3)
P fastmap 1.2.0 2024-05-15 [?] CRAN (R 4.4.0)
P forcats * 1.0.0 2023-01-29 [?] CRAN (R 4.3.0)
P foreach 1.5.2 2022-02-02 [?] CRAN (R 4.3.0)
P furrr 0.3.1 2022-08-15 [?] CRAN (R 4.3.0)
P future 1.33.2 2024-03-26 [?] CRAN (R 4.3.1)
P future.apply 1.11.2 2024-03-28 [?] CRAN (R 4.3.1)
P generics 0.1.3 2022-07-05 [?] CRAN (R 4.3.0)
P ggplot2 * 3.5.1 2024-04-23 [?] CRAN (R 4.3.1)
P globals 0.16.3 2024-03-08 [?] CRAN (R 4.3.1)
glue 1.8.0 2024-09-30 [1] RSPM (R 4.4.0)
P gower 1.0.1 2022-12-22 [?] CRAN (R 4.3.0)
P GPfit 1.0-8 2019-02-08 [?] CRAN (R 4.3.0)
P gtable 0.3.5 2024-04-22 [?] CRAN (R 4.3.1)
P hardhat 1.4.0 2024-06-02 [?] CRAN (R 4.4.0)
P here 1.0.1 2020-12-13 [?] CRAN (R 4.3.0)
P hms 1.1.3 2023-03-21 [?] CRAN (R 4.3.0)
P htmltools 0.5.8.1 2024-04-04 [?] CRAN (R 4.3.1)
P htmlwidgets 1.6.4 2023-12-06 [?] CRAN (R 4.3.1)
P infer * 1.0.7 2024-03-25 [?] CRAN (R 4.3.1)
P ipred 0.9-14 2023-03-09 [?] CRAN (R 4.3.0)
P iterators 1.0.14 2022-02-05 [?] CRAN (R 4.3.0)
P jsonlite 1.8.8 2023-12-04 [?] CRAN (R 4.3.1)
P knitr 1.47 2024-05-29 [?] CRAN (R 4.4.0)
P labeling 0.4.3 2023-08-29 [?] CRAN (R 4.3.0)
P lattice 0.22-6 2024-03-20 [?] CRAN (R 4.4.0)
P lava 1.8.0 2024-03-05 [?] CRAN (R 4.3.1)
P lhs 1.1.6 2022-12-17 [?] CRAN (R 4.3.0)
P lifecycle 1.0.4 2023-11-07 [?] CRAN (R 4.3.1)
P listenv 0.9.1 2024-01-29 [?] CRAN (R 4.3.1)
P lubridate * 1.9.3 2023-09-27 [?] CRAN (R 4.3.1)
P magrittr 2.0.3 2022-03-30 [?] CRAN (R 4.3.0)
P MASS 7.3-61 2024-06-13 [?] CRAN (R 4.4.0)
P Matrix 1.7-0 2024-03-22 [?] CRAN (R 4.4.0)
P modeldata * 1.4.0 2024-06-19 [?] CRAN (R 4.4.0)
P modelenv 0.1.1 2023-03-08 [?] CRAN (R 4.3.0)
P munsell 0.5.1 2024-04-01 [?] CRAN (R 4.3.1)
P nnet 7.3-19 2023-05-03 [?] CRAN (R 4.4.0)
P parallelly 1.37.1 2024-02-29 [?] CRAN (R 4.3.1)
P parsnip * 1.2.1 2024-03-22 [?] CRAN (R 4.3.1)
P pillar 1.9.0 2023-03-22 [?] CRAN (R 4.3.0)
P pkgconfig 2.0.3 2019-09-22 [?] CRAN (R 4.3.0)
P prodlim 2023.08.28 2023-08-28 [?] CRAN (R 4.3.0)
P purrr * 1.0.2 2023-08-10 [?] CRAN (R 4.3.0)
P R6 2.5.1 2021-08-19 [?] CRAN (R 4.3.0)
P ranger * 0.16.0 2023-11-12 [?] RSPM
P Rcpp 1.0.12 2024-01-09 [?] CRAN (R 4.3.1)
P readr * 2.1.5 2024-01-10 [?] CRAN (R 4.3.1)
P recipes * 1.0.10 2024-02-18 [?] CRAN (R 4.3.1)
renv 1.0.7 2024-04-11 [1] CRAN (R 4.4.0)
P rlang 1.1.4 2024-06-04 [?] CRAN (R 4.3.3)
P rmarkdown 2.27 2024-05-17 [?] CRAN (R 4.4.0)
P rpart * 4.1.23 2023-12-05 [?] CRAN (R 4.4.0)
P rprojroot 2.0.4 2023-11-05 [?] CRAN (R 4.3.1)
P rsample * 1.2.1 2024-03-25 [?] CRAN (R 4.3.1)
P rstudioapi 0.16.0 2024-03-24 [?] CRAN (R 4.3.1)
P scales * 1.3.0.9000 2024-05-07 [?] Github (r-lib/scales@c0f79d3)
P sessioninfo 1.2.2 2021-12-06 [?] CRAN (R 4.3.0)
P stringi 1.8.4 2024-05-06 [?] CRAN (R 4.3.1)
P stringr * 1.5.1 2023-11-14 [?] CRAN (R 4.3.1)
P survival 3.7-0 2024-06-05 [?] CRAN (R 4.4.0)
P tibble * 3.2.1 2023-03-20 [?] CRAN (R 4.3.0)
P tidymodels * 1.2.0 2024-03-25 [?] CRAN (R 4.3.1)
P tidyr * 1.3.1 2024-01-24 [?] CRAN (R 4.3.1)
P tidyselect 1.2.1 2024-03-11 [?] CRAN (R 4.3.1)
P tidyverse * 2.0.0 2023-02-22 [?] CRAN (R 4.3.0)
P timechange 0.3.0 2024-01-18 [?] CRAN (R 4.3.1)
P timeDate 4032.109 2023-12-14 [?] CRAN (R 4.3.1)
P tune * 1.2.1 2024-04-18 [?] CRAN (R 4.3.1)
P tzdb 0.4.0 2023-05-12 [?] CRAN (R 4.3.0)
P utf8 1.2.4 2023-10-22 [?] CRAN (R 4.3.1)
P vctrs 0.6.5 2023-12-01 [?] CRAN (R 4.3.1)
P vroom 1.6.5 2023-12-05 [?] CRAN (R 4.3.1)
withr 3.0.1 2024-07-31 [1] RSPM (R 4.4.0)
P workflows * 1.1.4 2024-02-19 [?] CRAN (R 4.3.1)
P workflowsets * 1.1.0 2024-03-21 [?] CRAN (R 4.3.1)
P xfun 0.45 2024-06-16 [?] CRAN (R 4.4.0)
P yaml 2.3.8 2023-12-11 [?] CRAN (R 4.3.1)
P yardstick * 1.3.1 2024-03-21 [?] CRAN (R 4.3.1)
[1] /Users/soltoffbc/Projects/info-5001/course-site/renv/library/macos/R-4.4/aarch64-apple-darwin20
[2] /Users/soltoffbc/Library/Caches/org.R-project.R/R/renv/sandbox/macos/R-4.4/aarch64-apple-darwin20/f7156815
P ── Loaded and on-disk path mismatch.
──────────────────────────────────────────────────────────────────────────────