usethis::use_git_config(
user.name = "Your name",
user.email = "Email associated with your GitHub account"
)Lab 00 - Hello R!
This lab is due September 2 September 3 at 11:59pm ET.
This lab will introduce you to the course computing workflow. The main goal is to reinforce our demo of R and RStudio, which we will be using throughout the course both to learn the statistical concepts discussed in the course and to analyze real data and come to informed conclusions.
R is the name of the programming language itself and RStudio is a convenient interface.
An additional goal is to reinforce Git and GitHub, the collaboration and version control system that we will be using throughout the course.
Git is a version control system (like “Track Changes” features from Microsoft Word but more powerful) and GitHub is the home for your Git-based projects on the internet (like DropBox but much better).
As the labs progress, you are encouraged to explore beyond what the labs dictate; a willingness to experiment will make you a much better programmer. Before we get to that stage, however, you need to build some basic fluency in R. Today we begin with the fundamental building blocks of R and RStudio: the interface, reading in data, and basic commands.
To make versioning simpler, this and the next lab are solo labs. In the future, you’ll learn about collaborating on GitHub and producing a single lab report for your lab team, but for now, concentrate on getting the basics down.
By the end of the lab, you will…
- Be familiar with the workflow using R, RStudio, Git, and GitHub
- Gain practice writing a reproducible report using Quarto
- Practice version control using GitHub
- Be able to create data visualizations using
ggplot2
Getting started
Your lab TAs will lead you through the Getting Started and Packages sections.
Access RStudio
If you plan to use your own computer
- Make sure you have completed the software setup instructions.
- Open RStudio.
If you plan to use RStudio Workbench
- Go to https://rstudio-workbench.infosci.cornell.edu and log in with your Cornell NetID and Password.
- Click the “New Session” button on the top of the page. Leave all the settings on their default state and click “Start Session”. You should now see an RStudio session.
If this is your first time accessing RStudio Workbench for the course, it will take a couple of minutes to prepare your session. Please be patient. When you start a session in the future, your container will already be prepared and it should start within 15 seconds.
Setup your GitHub authentication
Run the following code in the R console to ensure you have the required packages installed:
install.packages(c("usethis", "gitcreds", "gh"))In order to push changes to GitHub, you need to authenticate yourself. That is, you need to prove you are the owner of your GitHub account. When you log in to GitHub.com from your browser, you provide your username and password to prove your identity. But when you want to push and pull from your computer, you cannot use this method. Instead, you will prove your identity using one of two methods.
Authenticate using a Personal Access Token (PAT)
This method is preferred since it allows for seamless communication between R and Git for all possible applications.
A personal access token (or PAT) is a string of characters that can be used to authenticate a user when accessing a computer system instead of a username and password. Many online services are shifting towards requiring PATs for security reasons.
With this method you will clone repositories using a regular HTTPS url like https://github.com/<OWNER>/<REPO>.git.
Configure the Git credential helper by running the following R code in the console:
usethis::use_git_config(credential.helper = "store")Create your personal access token
Run this code from your R console:
usethis::create_github_token(
scopes = c("repo", "user", "gist", "workflow"),
description = "RStudio Workbench",
host = "https://github.coecis.cornell.edu/"
)This is a helper function that takes you to the web form to create a PAT.
- Give the PAT a description (e.g. “PAT for INFO 5001”)
- Leave the remaining options on the pre-filled form selected and click “Generate token”. As the page says, you must store this token somewhere, because you’ll never be able to see it again, once you leave that page or close the window. For now, you can copy it to your clipboard (we will save it in the next step).
If you lose or forget your PAT, just generate a new one.
Store your PAT
In order to store your PAT so you don’t have to reenter it every time you interact with Git, we need to run the following code:
gitcreds::gitcreds_set(url = "https://github.coecis.cornell.edu/")When prompted, paste your PAT into the console and press return. Your credential should now be saved on your computer.
Confirm your PAT is saved
Run the following code:
gh::gh_whoami(.api_url = "https://github.coecis.cornell.edu/")
usethis::git_sitrep()You should see output that provides information about your GitHub account.
Authenticate using Secure Shell Protocol
You can use this approach to authenticate yourself on GitHub. Note that you may find some limitations communicating with Git outside of standard processes (e.g. cloning/pushing/pulling repos directly), and will still need to create a PAT for some course assignments. However for students using RStudio Workbench, the SSH method will work for the entire semester (i.e. set it up once and never have to worry about it again).
The Secure Shell Protocol (SSH) is another method for authenticating your identity when communicating with GitHub. While a password can eventually be cracked with a brute force attack, SSH keys are nearly impossible to decipher by brute force alone. Generating a key pair provides you with two long strings of characters: a public and a private key. You can place the public key on any server (like GitHub), and then unlock it by connecting to it with a client that already has the private key (your computer or RStudio Serve). When the two match up, the system unlocks without the need for a password.
The URL for SSH remotes looks like git@github.com:<OWNER>/<REPO>.git. Make sure you use this URL to clone a repository. If you accidentally use the HTTPS version, the operation will not work.
Set up your SSH key
You only need to do this authentication process one time on a single system.
Type
credentials::ssh_keygen()into your console.R will ask “No SSH key found. Generate one now?” You should click 1 for yes.
-
You will generate a key. It will begin with “ssh-rsa….” and look something like this:
$key [1] "/home/bcs88/.ssh/id_rsa" $pubkey [1] "ssh-rsa AAAAB3NzaC1yc2EAAAADAQABAAABAQDJYmJeave083exQwYcIqZJk/Y1mgPxdcTYCTWLL+6mlhN9MM3enjDqb2eZvVJ0JK29NYL1++DTqY/saP08IlswNIMntwaWFDNx42yLsuFrWiPqzm9hWWnRcor/d+4zTrcSIEvfAAnLsYkagNqurrCf2taO62YRepTgxErLvLOG10qn4LKhNfT+PTqdPq2Mr88jXQYYrRxGnOV6oVYf6PurKkiooTsKYxVtJWai8Ek9fhK2y5vaQd5yP0H/3Hbw8Mn+rB+O8Yj6/oQKGBCgxkDB4Aw7T91DkIXlHppneO683Y54WvUftJYvSVsnyt/XuNjvXNAir0+kHETLM32uzH6L" Copy the entire string of characters (not including the quotation marks) and paste them into the settings page on GitHub. Give the key an informative title such as “INFO 5001 RStudio Workbench”. Click “Add SSH key.”
Configure Git
There is one more thing we need to do before getting started on the assignment. Specifically, we need to configure your git so that RStudio can communicate with GitHub. This requires two pieces of information: your name and email address.
To do so, you will use the use_git_config() function from the usethis package. Type the following lines of code in the console in RStudio filling in your name and the email address associated with your GitHub account.
For example, mine would be
usethis::use_git_config(
user.name = "Benjamin Soltoff",
user.email = "bcs88@cornell.edu"
)You are now ready interact with GitHub via RStudio!
Clone the repo & start new RStudio project
Go to the course organization at https://github.coecis.cornell.edu/info5001-fa24 organization on GitHub. Click on the repo with the prefix lab-00. It contains the starter documents you need to complete the lab.
Click on the green CODE button, select HTTPS or SSH based on the authentication method you set up previously. Click on the clipboard icon to copy the repo URL.
In RStudio, go to File ➛ New Project ➛ Version Control ➛ Git.
Copy and paste the URL of your assignment repo into the dialog box Repository URL.
Click Create Project, and the files from your GitHub repo will be displayed in the Files pane in RStudio.
Click lab-00-datasaurus.qmd to open the template Quarto file. This is where you will write up your code and narrative for the lab.
R and R Studio
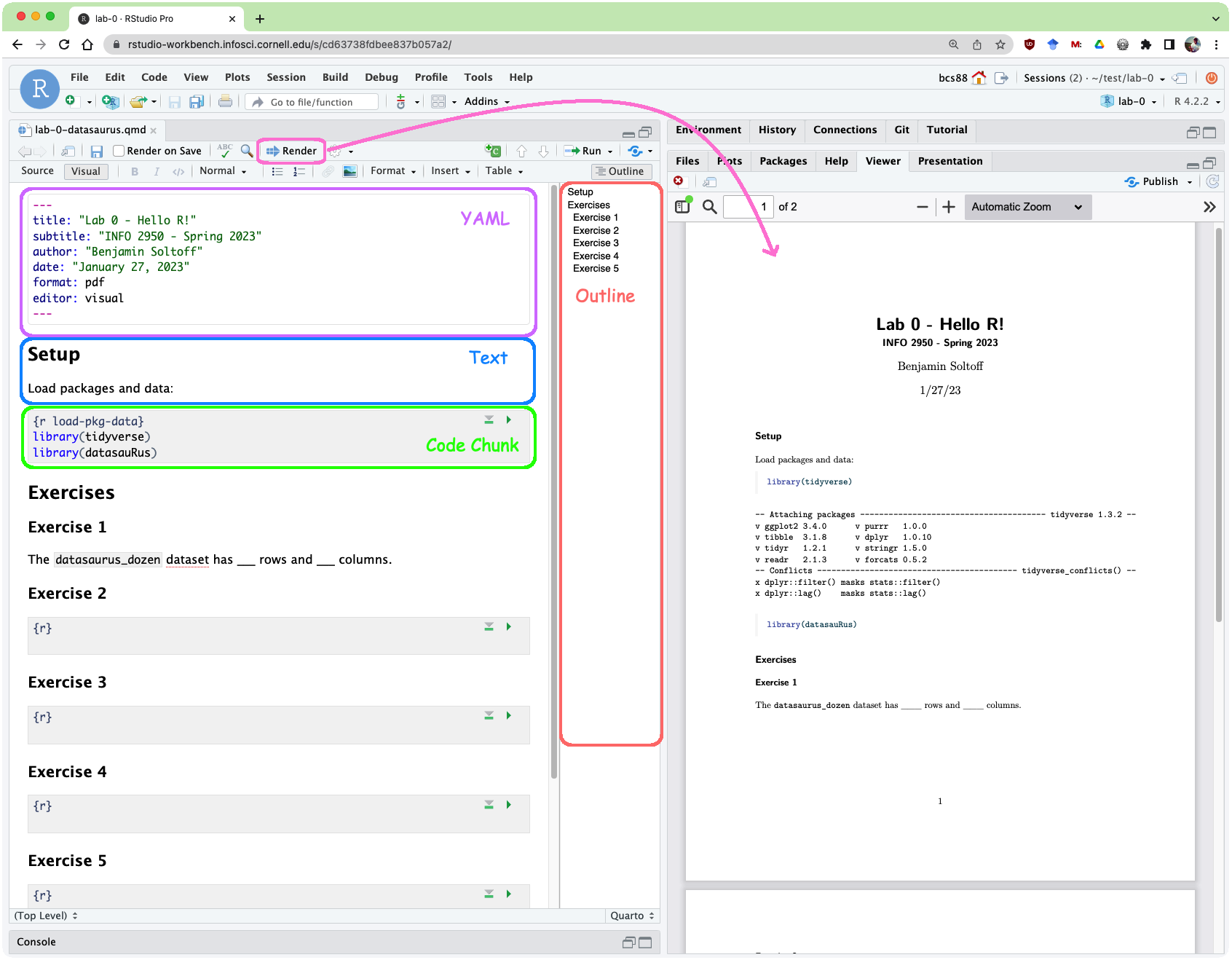
Below are the components of the RStudio IDE.
Below are the components of a Quarto (.qmd) file.
YAML
The top portion of your Quarto file (between the three dashed lines) is called YAML. It stands for “YAML Ain’t Markup Language”. It is a human friendly data serialization standard for all programming languages. All you need to know is that this area is called the YAML (we will refer to it as such) and that it contains meta information about your document.
Open the Quarto (.qmd) file in your project, change the author name to your name, and render the document. Examine the rendered document.
Committing changes
Now, go to the Git pane in your RStudio instance. This will be in the top right hand corner in a separate tab.
If you have made changes to your Quarto (.qmd) file, you should see it listed here. Click on it to select it in this list and then click on Diff. This shows you the difference between the last committed state of the document and its current state including changes. You should see deletions in red and additions in green.
If you’re happy with these changes, we’ll prepare the changes to be pushed to your remote repository. First, stage your changes by checking the appropriate box on the files you want to prepare. Next, write a meaningful commit message (for instance, “updated author name”) in the Commit message box. Finally, click Commit. Note that every commit needs to have a commit message associated with it.
You don’t have to commit after every change, as this would get quite tedious. You should commit states that are meaningful to you for inspection, comparison, or restoration.
In the first few assignments we will tell you exactly when to commit and in some cases, what commit message to use. As the semester progresses we will let you make these decisions.
Now let’s make sure all the changes went to GitHub. Go to your GitHub repo and refresh the page. You should see your commit message next to the updated files. If you see this, all your changes are on GitHub and you’re good to go!
Push changes
Now that you have made an update and committed this change, it’s time to push these changes to your repo on GitHub.
In order to push your changes to GitHub, you must have staged your commit to be pushed. Click on Push.
Packages
In this lab we will work with two packages:
- datasauRus which contains the dataset, and
- tidyverse which is a collection of packages for doing data analysis in a “tidy” way.
Render the document which loads these two packages with the library() function.
The rendered document will include a message about which packages the tidyverse package is loading along with it. It’s just R being informative, a message does not indicate anything is wrong (it’s not a warning or an error).
The tidyverse is a meta-package. When you load it you get eight packages loaded for you:
- ggplot2: for data visualization
- dplyr: for data wrangling
- tidyr: for data tidying and rectangling
- readr: for reading and writing data
- tibble: for modern, tidy data frames
- stringr: for string manipulation
- forcats: for dealing with factors
- lubridate: for dealing with dates/times
- purrr: for iteration with functional programming
The message that’s printed when you load the package tells you which versions of these packages are loaded as well as any conflicts they may have introduced, e.g., the filter() function from dplyr has now masked (overwritten) the filter() function available in base R (and that’s ok, we’ll use dplyr::filter() anyway).
You can now Render your template document and see the results.
Data
The data frame we will be working with today is called datasaurus_dozen and it’s in the datasauRus package. Actually, this single data frame contains 13 datasets, designed to show us why data visualization is important and how summary statistics alone can be misleading. The different datasets are marked by the dataset variable.
If it’s confusing that the data frame is called datasaurus_dozen when it contains 13 datasets, you’re not alone! Have you heard of a baker’s dozen?
Let’s also load these packages in the Console. You can do this by either typing the following in the console or clicking on the play button (green triangle) on the code chunk that loads the packages.
To find out more about the dataset, type the following in your console.
?datasaurus_dozenA question mark before the name of an object will always bring up its help file. This command must be run in the console. Alternatively, you can use the help() function.
help(datasaurus_dozen)Exercises
- Based on the help file, how many rows and how many columns does the
datasaurus_dozenfile have? What are the variables included in the data frame? Add your responses to your lab report.
When you’re done, commit your changes with the commit message “Added answer for Ex 1”,
Then, push these changes.
Let’s take a look at what these datasets are. To do so we can check the distinct() values of the dataset variable:
datasaurus_dozen |>
distinct(dataset)# A tibble: 13 × 1
dataset
<chr>
1 dino
2 away
3 h_lines
4 v_lines
5 x_shape
6 star
7 high_lines
8 dots
9 circle
10 bullseye
11 slant_up
12 slant_down
13 wide_linesThe original Datasaurus (dino) was created by Alberto Cairo in this great blog post. The other Dozen were generated using simulated annealing and the process is described in the paper Same Stats, Different Graphs: Generating Datasets with Varied Appearance and Identical Statistics through Simulated Annealing by Justin Matejka and George Fitzmaurice.1 In the paper, the authors simulate a variety of datasets that the same summary statistics to the Datasaurus but have very different distributions.
1 Matejka, Justin, and George Fitzmaurice. “Same stats, different graphs: Generating datasets with varied appearance and identical statistics through simulated annealing.” Proceedings of the 2017 CHI Conference on Human Factors in Computing Systems. ACM, 2017.
Data visualization and summary
- Plot
xvs.yfor thedinodataset. Then, calculate the correlation coefficient betweenxandyfor this dataset.
Below is the code you will need to complete this exercise. Basically, the answer is already given, but you need to include relevant bits in your document and successfully render it and view the results.
Start with the datasaurus_dozen and pipe it into the filter function to filter for observations where dataset == "dino". Store the resulting filtered data frame as a new data frame called dino_data.
dino_data <- datasaurus_dozen |>
filter(dataset == "dino")There is a lot going on here, so let’s slow down and unpack it a bit.
First, the pipe operator: |>, takes what comes before it and sends it as the first argument to what comes after it. So here, we’re saying filter the datasaurus_dozen data frame for observations where dataset == "dino".
Second, the assignment operator: <-, assigns the name dino_data to the filtered data frame.
Next, we need to visualize these data. We will use the ggplot function for this. Its first argument is the data you’re visualizing. Next we define the aesthetic mappings. In other words, the columns of the data that get mapped to certain aesthetic features of the plot, e.g. the x axis will represent the variable called x and the y axis will represent the variable called y. Then, we add another layer to this plot where we define which geometric shapes we want to use to represent each observation in the data. In this case we want these to be points, hence geom_point.
ggplot(data = dino_data, mapping = aes(x = x, y = y)) +
geom_point()For the second part of this exercise, we need to calculate a summary statistic: the correlation coefficient. Correlation coefficient, often referred to as \(r\) in statistics, measures the linear association between two variables. You will see that some of the pairs of variables we plot do not have a linear relationship between them. This is exactly why we want to visualize first: visualize to assess the form of the relationship, and calculate \(r\) only if relevant. In this case, calculating a correlation coefficient really doesn’t make sense since the relationship between x and y is definitely not linear (it’s dinosaurial)!
For illustrative purposes only, let’s calculate the correlation coefficient between x and y.
Start with dino_data and calculate a summary statistic that we will call r as the correlation between x and y.
This is a good place to pause, render, and commit changes with the commit message “Added answer for Ex 2.”
Then, push these changes when you’re done.
- Plot
xvs.yfor thecircledataset. You can (and should) reuse code we introduced above, just replace the dataset name with the desired dataset. Then, calculate the correlation coefficient betweenxandyfor this dataset. How does this value compare to therofdino?
This is another good place to pause, render, and commit changes with the commit message “Added answer for Ex 3.”
Then, push these changes when you’re done.
- Plot
xvs.yfor thestardataset. You can (and should) reuse code we introduced above, just replace the dataset name with the desired dataset. Then, calculate the correlation coefficient betweenxandyfor this dataset. How does this value compare to therofdino?
You should pause again, render, commit changes with the commit message “Added answer for Ex 4”.
Then, push.
- Finally, let’s plot all datasets at once. In order to do this we will make use of faceting, given by the code below:
Facet by the dataset variable, placing the plots in a 3 column grid, and don’t add a legend.
ggplot(datasaurus_dozen, aes(x = x, y = y, color = dataset)) +
geom_point() +
facet_wrap(facets = vars(dataset), ncol = 3) +
theme(legend.position = "none")And we can use the group_by function to generate all the summary correlation coefficients. We’ll go through these functions in a couple of weeks when we learn about data wrangling.
Include the faceted plot and the summary of the correlation coefficients in your lab write-up by including relevant code in R chunks (and give them appropriate labels). In the narrative below the code chunks, briefly comment on what you notice about the plots and the correlations between x and y values within each of them (one or two sentences is fine!).
You should pause again, render, commit changes with the commit message “Added answer for Ex 5”.
Then, push.
You’re done with the data analysis exercises, but we’d like to do one more thing to customize the look of the report.
Resize your figures
We can customize the output from a particular R chunk by including options in the header that will override any global settings.
- In the R chunks you wrote for Exercises 2-5, customize the settings by modifying the options in the R chunks used to create those figures.
For Exercises 2, 3, and 4, we want square figures. We can use fig-height and fig-width in the options to adjust the height and width of figures. Modify the chunks in Exercises 2-4 to be as follows:
```{r}
#| label: ex2-chunk-label
#| fig-height: 5
#| fig-width: 5
# Your code that created the figure
```Code chunk labels need to be unique or you will get an error when you render the document. Make sure to fix the label in each chunk to be unique to the specific exercise. Even better, choose something substantive yet simple that describes the chunk rather than ex2-chunk-label.
For Exercise 5, modify your figure to have fig-height of 10 and fig-width of 6.
Now, save and render.
Once you’ve created this PDF file, you’re done!
Commit all remaining changes with the commit message “Done with Lab 0!”.
Then push.
Submission
Once you are finished with the lab, you will submit your final PDF document to Gradescope.
Before you wrap up the assignment, make sure all documents are updated on your GitHub repo. We will be checking these to make sure you have been practicing how to commit and push changes.
You must turn in a PDF file to the Gradescope page by the submission deadline to be considered “on time”.
To submit your assignment:
- Go to http://www.gradescope.com and click Log in in the top right corner.
- Click School Credentials \(\rightarrow\) Cornell University NetID and log in using your NetID credentials.
- Click on your INFO 5001 course.
- Click on the assignment, and you’ll be prompted to submit it.
- Mark all the pages associated with exercise. All the pages of your lab should be associated with at least one question (i.e., should be “checked”).
- Select all pages of your .pdf submission to be associated with Exercise 6.
- Select all pages of your .pdf submission to be associated with the “Workflow & formatting” question.
Grading (50 pts)
| Component | Points |
|---|---|
| Ex 1 | 5 |
| Ex 2 | 10 |
| Ex 3 | 5 |
| Ex 4 | 5 |
| Ex 5 | 10 |
| Ex 6 | 7 |
| Workflow & formatting | 8 |
The “Workflow & formatting” component assesses the reproducible workflow. This includes:
- Following tidyverse code style
- All code being visible in rendered PDF (no more than 80 characters)
- Appropriate figure sizing, and figures with informative labels and legends
Acknowledgments
- This assignment is derived from STA 199: Introduction to Data Science